aiQSAR
October 20, 2020
Collection of tools by Jadavpur University
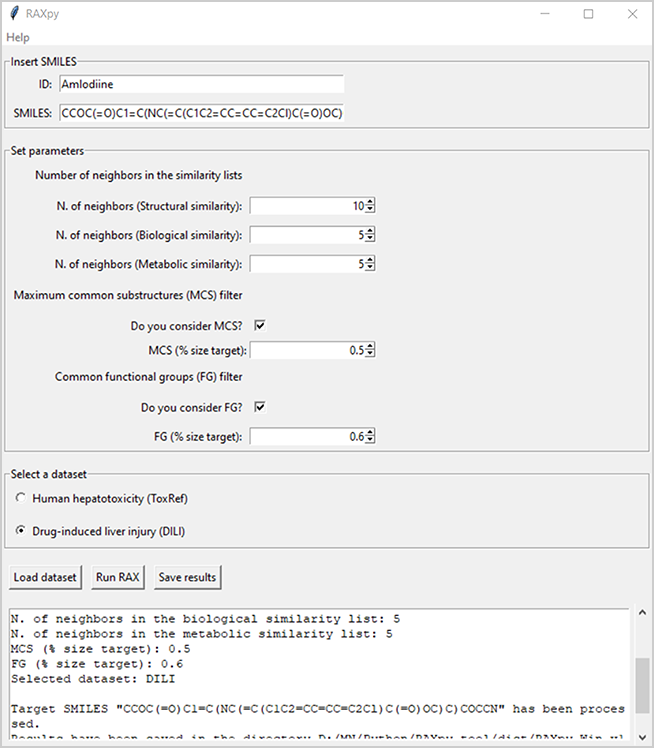
October 2, 2019RAXpy is a read-across tool based on structural, biological and metabolic similarities.
RAXpy is a read-across tool based on structural, biological and metabolic similarities. Filters based on the presence of maximum common substructures and common functional groups between the target and the source dataset can be applied to narrow the chemical space for the analogue(s) search.
RAXpy tool has been developed within the European Union's Horizon 2020 research and innovation programme under Grant Agreement No. 681002 (EU-ToxRisk).
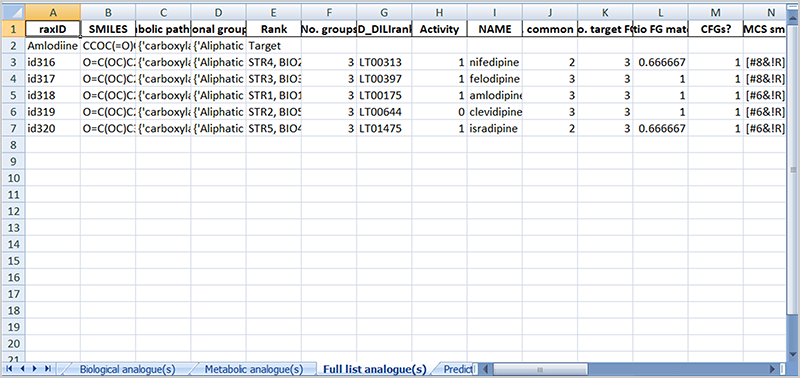
RAXpy is a read-across tool based on structural, biological and metabolic similarities. It allows to calculate similarities between a target molecule and chemicals from a source dataset. Two datasets are included related to high-tier in vivo toxicological endpoints, i.e., human hepatotoxicity (ToxRef) and drug induced liver injury (DILI). The predicted activity for the target is obtained using the activities of analogue(s) included in the similarity lists. Moreover, filters based on the presence of maximum common substructures and common functional groups between the target and the source compounds can be applied to narrow the chemical space for the analogue(s) search.
RAXpy tool is freely available for download through the repository hosted on Bitbucket
The software is a free and open-source application (GUI). Just download the tool, unpack the zipped file according to your system and run the executable file RAXpy:
- RAXpy Win v1.0.0 (64bit).zip (Windows)
- RAXpy Linux v1.0.0 (64bit).tar.gz (Linux)
RAXpy Screenshots
Developed by