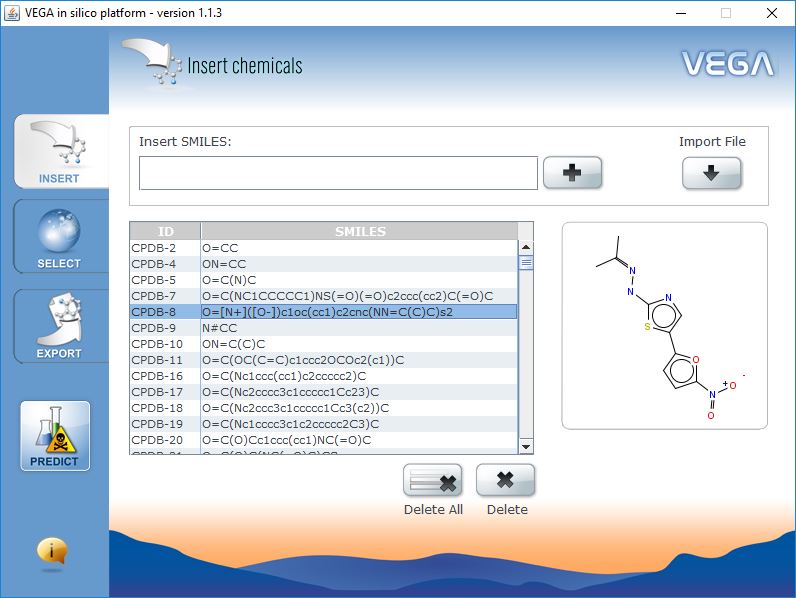
Input Data
VEGA QSAR works in batch mode. You can process thousands of molecules at the same time. You can decide to insert single SMILES manually, import a list of SMILES from text file, or import a SDF file.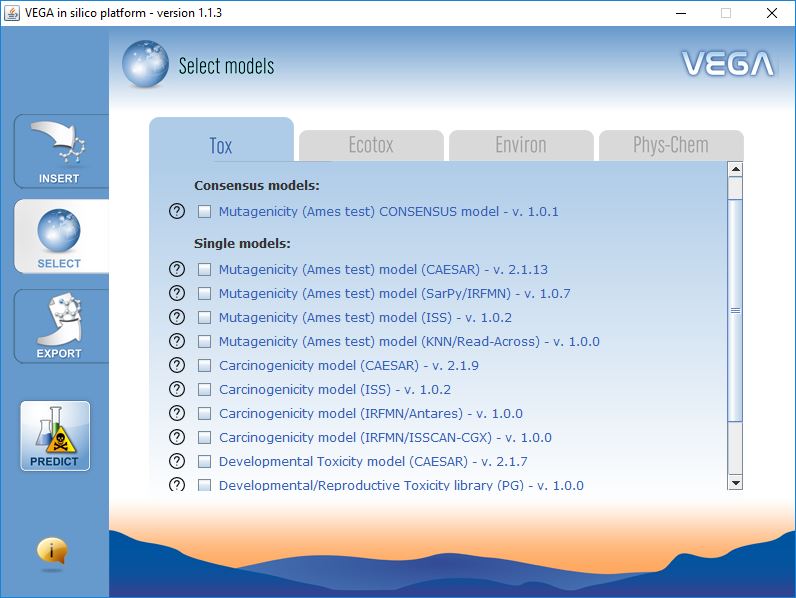
Model Selection
VEGA QSAR includes models for several endpoints, which have been organized in four classes: toxicity endpoints (e.g. mutagenicity, carcinogenicity, etc.), ecotoxicity endpoints (e.g. fish toxicity, bee toxicity, etc.), environmental-related properties (e.g. bioconcentration factor, persistence, etc.), physicochemical properties (e.g. LogP).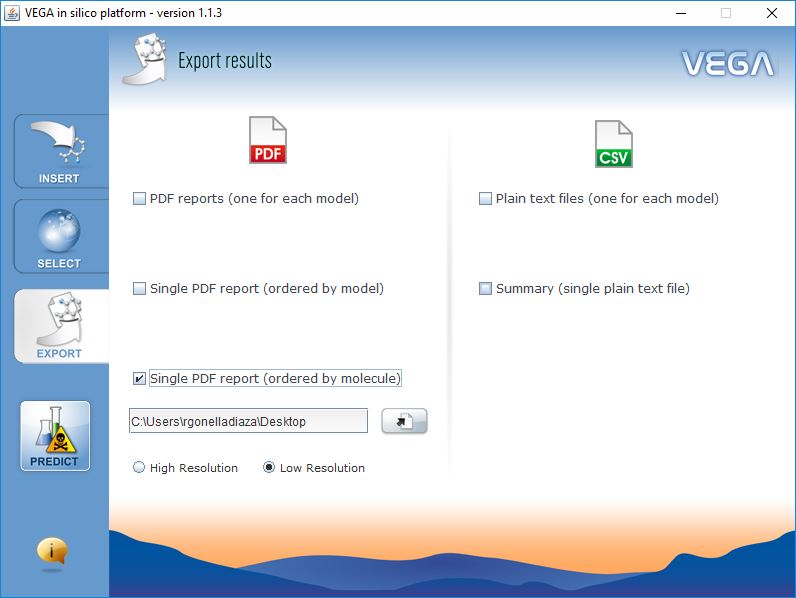
Export Results
VEGA QSAR can generate two type of reports:
- PDF Reports, which include the most complete information, similar molecules, reasoning on structural alerts, etc.
- CSV Reports, which are simple comma-separated text files, are useful when you need to predict large dataset of chemicals.
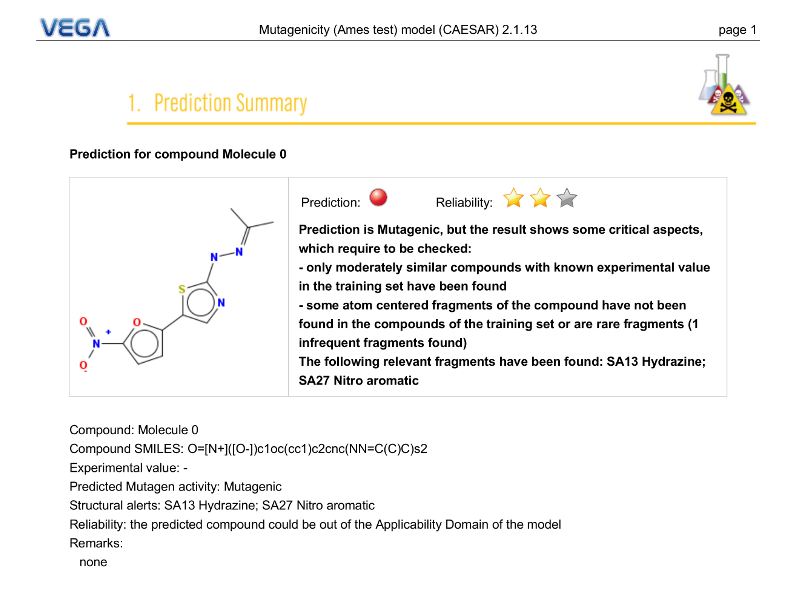
Prediction Summary
PDF Report
The first page gives an overview of the evaluation of the selected model for the target molecule. You can find information about the prediction, the applicability domain (reliability), reasoning, etc.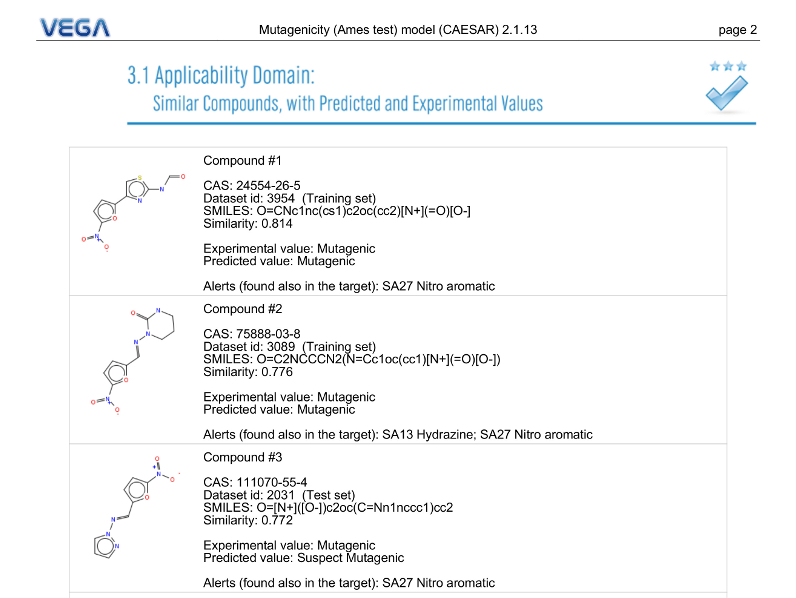
Similar Compounds
PDF Report
VEGA QSAR compare the target molecules with its internal dataset and lists the six most similar molecules for which it has experimental value information. For each similar compound, VEGA QSAR provide the experimental value, the prediction, a 2D structure representation, structural alerts in common with the target molecules (etc.)
Applicability Domain
PDF Report
To evaluate the reliability of the prediction obtained, VEGA QSAR has been provided with an applicability domain evaluation tool. VEGA QSAR, using several parameters, compare the target molecule with the model training set in order to check if it falls within the applicability domain of the model.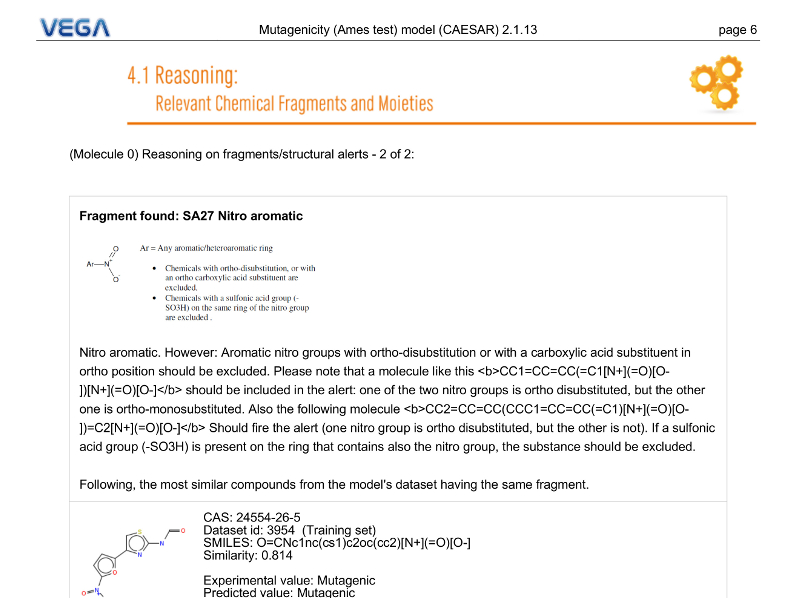
Reasoning
PDF Report
Depending on model, VEGA QSAR can provided different type of reasoning regarding the prediction. In this example the software informs the user about the presence of a mutagenic-related structural alert in the target molecule.
VEGA Screenshots
